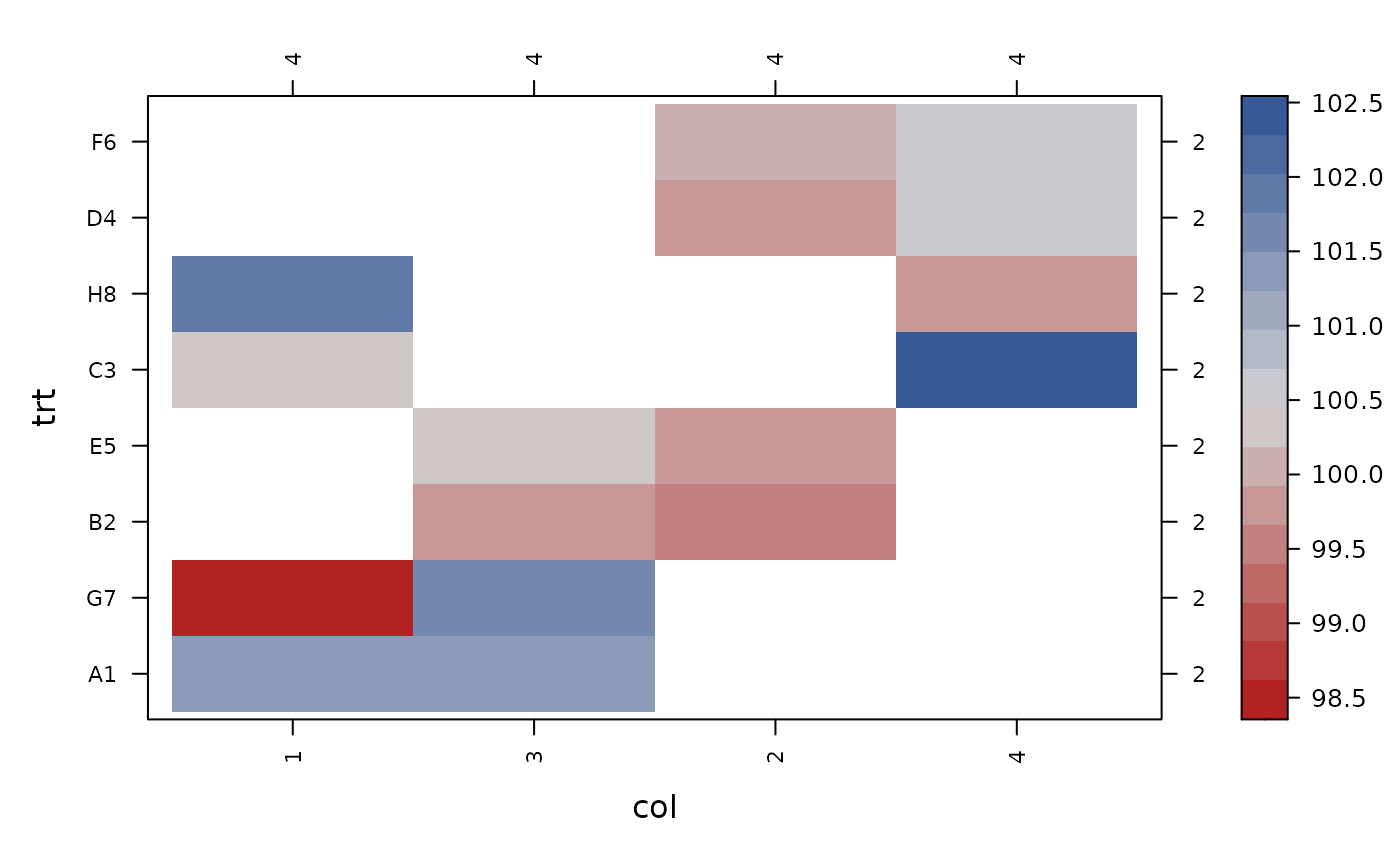Data from Eccleston & Russell
Source
Eccleston, J. and K. Russell (1975). Connectedness and orthogonality in multi-factor designs. Biometrika, 62, 341-345. https://doi.org/10.1093/biomet/62.2.341
Details
A dataframe with 3 treatment factors. Each pair of factors is connected, but the 3 factors are disconnected. The 'trt' column uses numbers to match Eccleston (Table 1, Design 1) and letters to match Foulley (Table 13.3).
References
Foulley, J. L., Bouix, J., Goffinet, B., & Elsen, J. M. (1990). Connectedness in Genetic Evaluation. Advanced Series in Agricultural Sciences, 277–308. https://doi.org/10.1007/978-3-642-74487-7_13
Examples
# Each pair of factors is connected
con_check(data_eccleston, ~ row + trt)
#> [1] 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1
#> Levels: 1
con_check(data_eccleston, ~ col + trt)
#> [1] 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1
#> Levels: 1
con_check(data_eccleston, ~ row + col)
#> [1] 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1
#> Levels: 1
# But all three factors are COMPLETELY disconnected
con_check(data_eccleston, ~ row + col + trt)
#> [1] 16 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15
#> Levels: 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16
set.seed(42)
data_eccleston <- transform(data_eccleston,
y = rnorm(nrow(data_eccleston), mean=100))
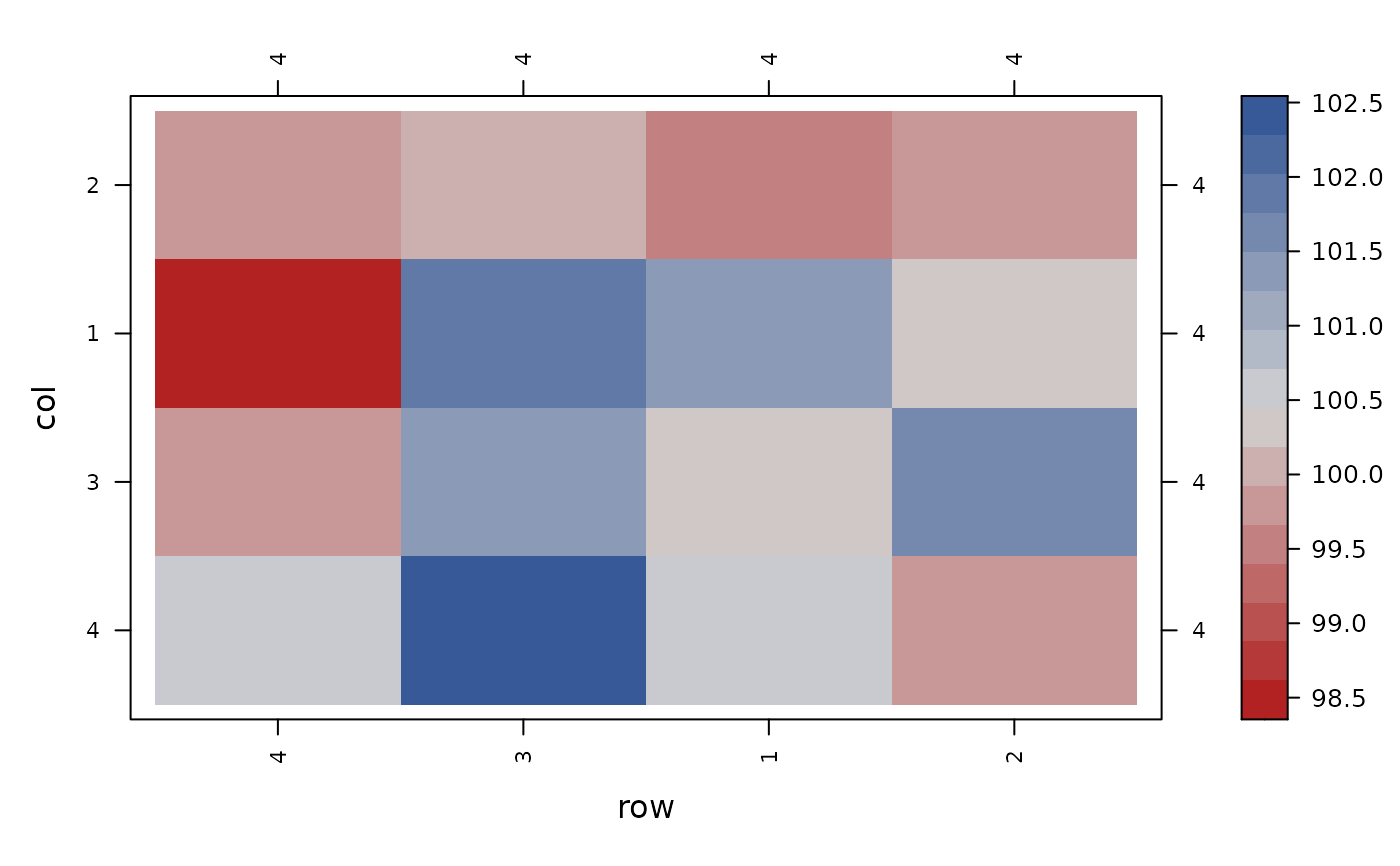
con_view(data_eccleston, y ~ row*col, xlab="row", ylab="col")
 con_view(data_eccleston, y ~ row*trt, xlab="row", ylab="trt")
con_view(data_eccleston, y ~ row*trt, xlab="row", ylab="trt")
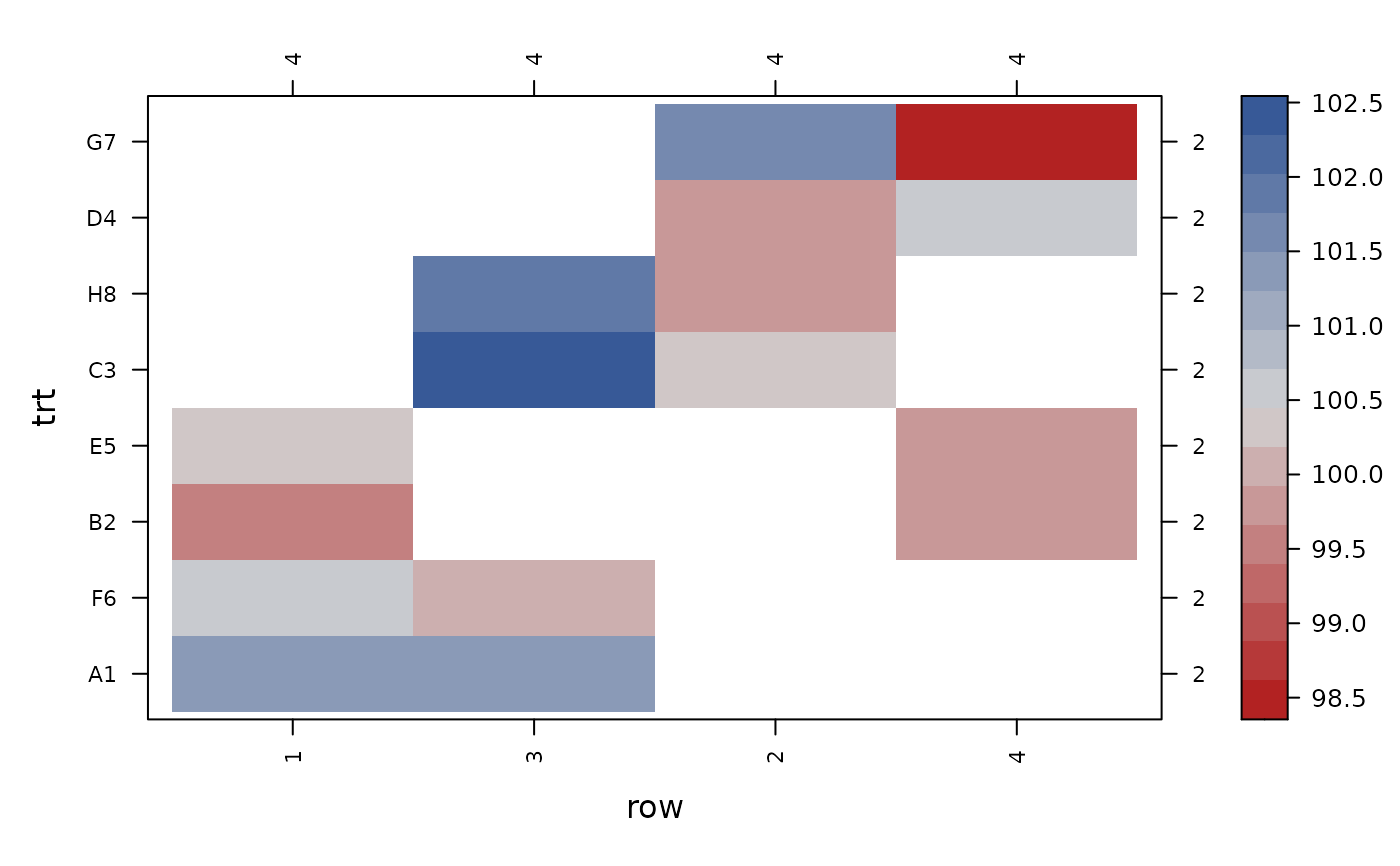 con_view(data_eccleston, y ~ col*trt, xlab="col", ylab="trt")
con_view(data_eccleston, y ~ col*trt, xlab="col", ylab="trt")