The NIPALS algorithm
Kevin Wright
2017-10-27
Source:vignettes/nipals_algorithm.Rmd
nipals_algorithm.RmdThe principal components of $\bf X'X$, where $\bf X$ is a column-centered matrix, can be found by several methods, including SVD and NIPALS.
Singular value decomposition
The SVD (Singular Value Decomposition) of a matrix is $$ \bf X = U S V', $$ where $\bf U$ and $\bf V$ are orthogonal matrices and $\bf S$ is a diagonal matrix of singular values.
SVD does not allow missing values in the data.
NIPALS
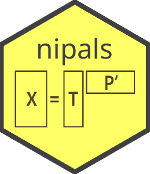
The NIPALS (Nonlinear Iterative Partial Least Squares) algorithm can be used to find the first few (or all) principal components with the decomposition $$ \bf X = \bf T \bf P ' $$ where the columns of $\bf T$ are called scores and the columns of $\bf P$ (the rows of $\bf P'$) are called the loadings.
The algorithm begins by initializing and $\bf X_h = \bf X$, then proceeds through the following basic steps:
- Choose $\bf t_h$ as any column of $\bf X_h$.
- Compute loadings $\bf p_h = X_h' t_h / t_h' t_h$.
- Let $\bf p_h = p_h / \sqrt{p_h' p_h}$.
- Compute scores $\bf t_h = X_h p_h / p_h' p_h$.
Repeat (3) and (4) until convergence for the principal component.
Let $\bf X_{h+1} = \bf X_h - t_h p_h'$. Let $\lambda_h = \bf t_h' t$ (eigen value). Increment and repeat for the next principal component.
Assemble the columns of $\bf T$ from the $\bf t_h$ and the columns of $\bf P$ from the vectors $\bf p_h$.
The resulting PCs may be scaled in different ways. One way to scale the PCA solution is to define the loadings $\bf P = V$ and $\bf T = U'S$.
Missing data
The NIPALS algorithm can be modified to accommodate missing values using the method of Martens and Martens (2001) (p. 381).
If, for a certain variable [column of $\bf X$], a missing value is encountered in $\bf X$ for a certain object [row of $\bf X$], then the corresponding elements in $\bf t_{ih}$ must also be skipped in the calculation of the loadings, which for $\bf X$-variable is $$ \bf p_{hk} = X_{k,h-1} t_h' / (t_h' t_h) . $$
Likewise, if, for a certain sample [row of $\bf X$], a missing value is encountered in $\bf X$ for a certain variable [column of $\bf X$], then the corresponding elements in $\bf p_{kh}$ must also be skipped in calculating the scores, which for sample is $$ \bf t_{ih} = X_{i,h-1} p_h / (p_h' p_h) $$ This method may have convergence problems if there are many missing values.
Gram-Schmidt orthogonalization
Because of the accumulation of floating-point errors, the orthogonality of the principal components is quickly lost as the number of components increases. Andrecut (2009) provided a Gram-Schmidt modified version of NIPALS that stabilizes the orthogonality by re-orthogonalizing the scores and loadings at each iteration. The ‘corrected’ terms are:
$$ \bf p_c = p - P_{1:h} P_{1:h}' p $$
and
$$ \bf t_c = t - T_{1:h} T_{1:h}' t $$ where $\bf P_{1:h}$ and $\bf T_{1:h}$ are the loadings and scores matrices based on the first principal components. Since $\bf P_{1:h} P_{1:h}'$ only needs to be calculated once for each PC (and incrementally), the orthogonalization is not very computationally expensive.
This correction method is also used by SAS PROC HPPRINCOMP (which does not allow missing values).
Example 1
A small dataset with two missing values.
## Loading required package: nipals
B <- matrix(c(50, 67, 90, 98, 120,
55, 71, 93, 102, 129,
65, 76, 95, 105, 134,
50, 80, 102, 130, 138,
60, 82, 97, 135, 151,
65, 89, 106, 137, 153,
75, 95, 117, 133, 155), ncol=5, byrow=TRUE)
B2 <- B
B2[1,1] <- B2[2,1] <- NA
m0 <- svd(scale(B)) # center and scale
require("nipals")
m1 <- nipals::nipals(B2, gramschmidt=FALSE)
m2 <- nipals::nipals(B2, gramschmidt=TRUE)Model m1 omits the Gram-Schmidt orthogonalization step
at each iteration. Model m2 includes it.
The eigenvalues for the two models are very similar.
round( m1$eig, 3)## [1] 4.876 2.044 1.073 0.237 0.143
round( m2$eig, 3)## [1] 4.876 2.035 1.079 0.234 0.133In theory, the loadings matrix $\bf P$ is orthogonal so that $\bf P' P = I$. If there are missing values, however, then the calculation of approximate PCs causes numerical errors to accumulate, so that in practice only the first few components can be accurately calculated. (The coordinates of the last PC can often be quite poor.)
In this small example, the first 3 PCs of model m1 are
fairly orthogonal, but the 4th and 5th PC are not so good. For model
m2, the PCs are nearly exactly orthogonal.
## PC1 PC2 PC3 PC4 PC5
## PC1 1.000 0.011 0.006 -0.043 -0.416
## PC2 0.011 1.000 -0.011 -0.003 0.125
## PC3 0.006 -0.011 1.000 -0.050 -0.058
## PC4 -0.043 -0.003 -0.050 1.000 -0.006
## PC5 -0.416 0.125 -0.058 -0.006 1.000## PC1 PC2 PC3 PC4 PC5
## PC1 1 0 0 0 0
## PC2 0 1 0 0 0
## PC3 0 0 1 0 0
## PC4 0 0 0 1 0
## PC5 0 0 0 0 1Also in theory, $\bf T' T = I$ (if eigenvalues are removed from T), but missing values again invalidate this identity, unless the Gram-Schmidt method is used.
## PC1 PC2 PC3 PC4 PC5
## PC1 1.000 -0.086 0.058 -0.287 0.218
## PC2 -0.086 1.000 0.003 -0.014 0.003
## PC3 0.058 0.003 1.000 0.013 -0.002
## PC4 -0.287 -0.014 0.013 1.000 0.004
## PC5 0.218 0.003 -0.002 0.004 1.000## PC1 PC2 PC3 PC4 PC5
## PC1 1 0 0 0 0
## PC2 0 1 0 0 0
## PC3 0 0 1 0 0
## PC4 0 0 0 1 0
## PC5 0 0 0 0 1