Hypoxia Data
Giorgi Chighladze
2019-09-13
hypoxia.RmdData do not always come in a format that you can readily use for analysis. Some cleaning and restructuring of data is often needed despite of the size of the dataset.
Raw data
This data was downloaded 15 Apr 2019 from TidyTuesday. The full description of the data are here: https://github.com/rfordatascience/tidytuesday/files/2343596/Hypoxia.Article.proof.pdf.
Briefly, the data:
X1 - Dummy. Row number.
Altitude - Altitude.
Air press - Air pressure.
ppO2 - Part of pressure related to oxygen.
Alv pO2 - Pressure of O2 in lungs.
% sat O2 - Percentage of hemoglobin molecules with O2.
Alv pCO2 - Pressure of carbion dioxide in lungs.
Alv pO2 with O2=100% - Maximum O2 pressure in lungs without mask.
% sat O2_1 - Best saturation possible without mask.
Alv pCO2_1 - Pressure of CO2 in lungs with 100% O2.Before looking at the data, make sure that we do not missinterpret any entry when reading the file. For csv and txt files it is safe to read each columns as text. Also, it is good practice to read only the first few rows (5 in this case) of data for this purpose.
library(tidyverse)
datfile <- system.file("messydata", "hypoxia.csv", package="untidydata2")
read_csv(datfile, col_types = cols(.default = "c"), n_max = 5) %>%
knitr::kable()| X1 | Altitude | Air press | ppO2 | Alv pO2 | % sat O2 | Alv pCO2 | Alv pO2 with O2=100% | % sat O2_1 | Alv pCO2_1 |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Ft/m | mmHg | Air=21% | mmHg on air | >90% desired | >35 best | NA | 100% O2 | 100% O2 |
| 2 | Sea level | 760 | 159 | 104 | 97 | 40 | 673 | 100 | 40 |
| 3 | 10k/3k | 523 | 110 | 67 | 90 | 36 | 436 | 100 | 40 |
| 4 | 20k/6.1k | 349 | 73 | 40 | 73 | 24 | 262 | 100 | 40 |
| 5 | 30k/9.1k | 226 | 47 | 18 | 24 | 24 | 139 | 99 | 40 |
Problems
There are several problems that one can note at first glance.
- variable description is included in the first row
- column names contain spaces and symbols
- first column is redandant and inaccurate (first row is assigned 1, even though it does not represent an observation)
- variable ‘Altitude’ contains two values in different units and non-numeric record (‘Sea level’)
- last three variables are the same as previous 3 columns, but at different oxygen content (percentge)
Tidying the data
First we need to fix the variable names. The best way is to manually define each variable name after reading the data without column names. Since the last 6 columns represent the same variable measured at different oxygen levels, we will incorporate this piece of info in the variable names (and use later when reshaping the data).
data <- read_csv(datfile,
col_types = cols(.default = "?"),
col_names = FALSE,
skip = 2) %>%
# remove first cloumn
select(-1) %>%
# set names of variables
set_names(c('altitude', 'air_pressure', 'ppO2',
'alveoli_pO2-21', 'sturation_O2-21', 'alveoli_pCO2-21',
'alveoli_pO2-100', 'sturation_O2-100', 'alveoli_pCO2-100'))After reading the data we will address remaining problems listed as follow:
tidy_data <- data %>%
# set 'Sea level' as 0 ft and 0 m in format matching the other records in altitude
mutate(altitude = ifelse(altitude == 'Sea level', '0k/0k', altitude)) %>%
# split altitude data into columns corresponding to each unit of measurement
separate(altitude, into = c('altitude_ft', 'altitude_m'), sep = '/') %>%
# convert altitude into numeric data by taking into account k
mutate_at(vars(starts_with('altitude')), parse_number) %>%
mutate_at(vars(starts_with('altitude')), funs(. * 1000)) %>%
# handle last 6 columns
gather(key, value, 5:10) %>%
# extract oxygen level (that was incorporated into the variable name earlier)
separate(key, into = c('variable', 'O2'), sep = '-', convert = TRUE) %>%
spread(variable, value) %>%
arrange(O2, altitude_ft)## Warning: funs() is soft deprecated as of dplyr 0.8.0
## Please use a list of either functions or lambdas:
##
## # Simple named list:
## list(mean = mean, median = median)
##
## # Auto named with `tibble::lst()`:
## tibble::lst(mean, median)
##
## # Using lambdas
## list(~ mean(., trim = .2), ~ median(., na.rm = TRUE))
## This warning is displayed once per session.| altitude_ft | altitude_m | air_pressure | ppO2 | O2 | alveoli_pCO2 | alveoli_pO2 | sturation_O2 |
|---|---|---|---|---|---|---|---|
| 0 | 0 | 760 | 159 | 21 | 40 | 104 | 97 |
| 10000 | 3000 | 523 | 110 | 21 | 36 | 67 | 90 |
| 20000 | 6100 | 349 | 73 | 21 | 24 | 40 | 73 |
| 30000 | 9100 | 226 | 47 | 21 | 24 | 18 | 24 |
| 40000 | 12000 | 141 | 29 | 21 | NaN | NaN | NaN |
| 50000 | 15200 | 87 | 18 | 21 | NaN | NaN | NaN |
| 0 | 0 | 760 | 159 | 100 | 40 | 673 | 100 |
| 10000 | 3000 | 523 | 110 | 100 | 40 | 436 | 100 |
| 20000 | 6100 | 349 | 73 | 100 | 40 | 262 | 100 |
| 30000 | 9100 | 226 | 47 | 100 | 40 | 139 | 99 |
| 40000 | 12000 | 141 | 29 | 100 | 36 | 58 | 84 |
| 50000 | 15200 | 87 | 18 | 100 | 24 | 16 | 15 |
Plot
We can plot some data.
tidy_data %>%
mutate(O2 = factor(O2, levels = c(21, 100), labels = c('21%', '100%'))) %>%
ggplot(aes(x = alveoli_pO2, y = altitude_m)) +
geom_point(aes(colour = O2), na.rm = TRUE) +
theme_light()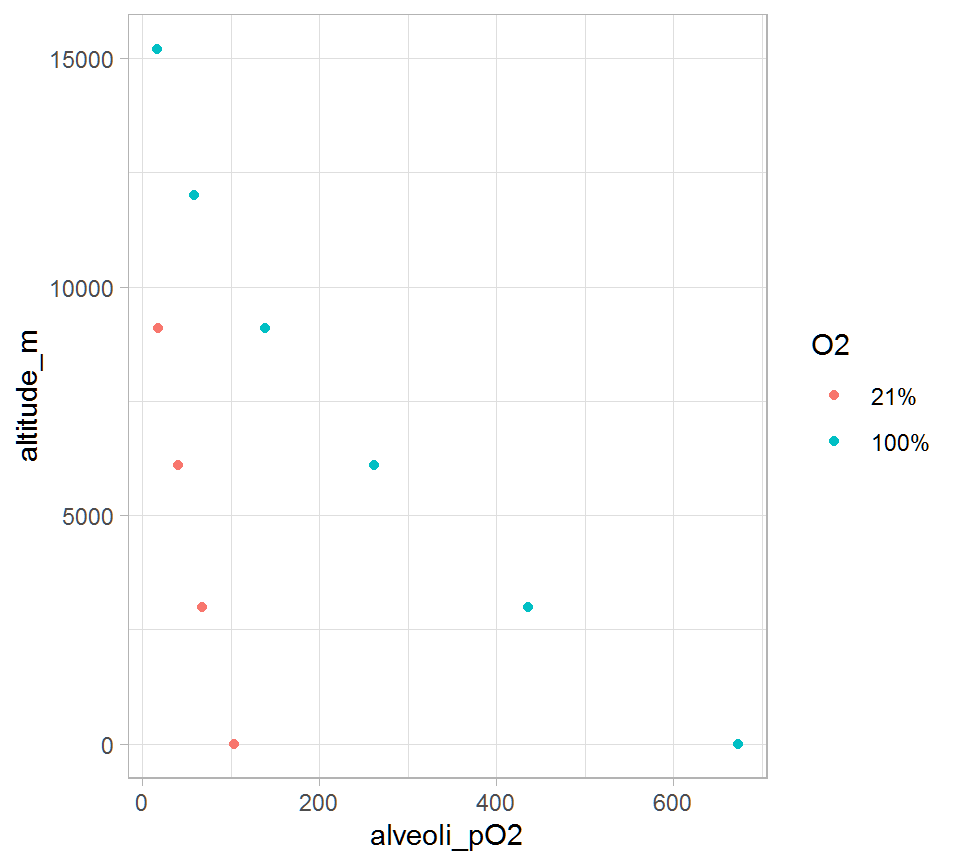
As an airplane pilot flies at higher altitudes, the pressure of oxygen in the lungs decreases.